Bioblitz 2016 is fast approaching. Even faster for those of us on the microbial team because we have to sample at least one month in advance in order to have data for the day of the Bioblitz. I’ll write up how/where/why we sample in the future, but today I’m going to go over how we can tell what microbes are present in a sample.
Once we have a sample in hand, the work can really begin.

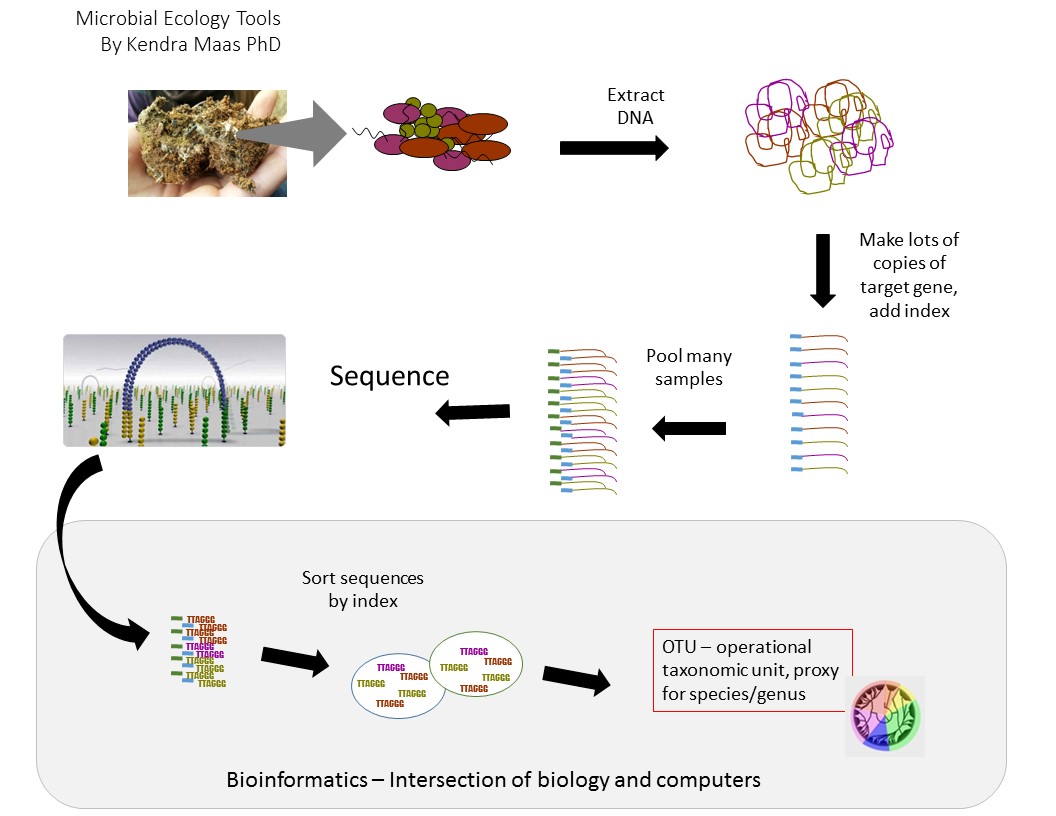
Within this soil sample is a huge number of organisms-plant roots, worms, nematodes, fungi, and of course lots and lots of bacteria. There are roughly 108 bacterial cells per gram of soil-100 MILLION cells in a chunk of soil that’s about as big as a lima bean. When we consider how we are going to figure out what bacteria make up those millions of cells, I find it useful to imagine just a few of them and follow the fate of those cells (and their DNA) through the process.
Imagine with me that these are the cells in that gram of soil.
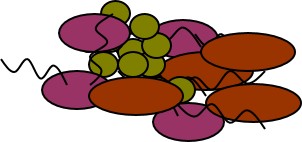
During the Bioblitz we will have microscopes set up so you can see bacteria from samples but it’s not possible to determine the species of bacteria based just on its appearance. One way to determine bacterial species is to look at the DNA sequence. To look at the DNA sequences, we have to get the DNA out of the cells and away from the rest of the sample. We will be using a high throughput DNA extraction kit provided by MoBio.
After extraction, we will have a mix of DNA from all the organisms in that soil sample.
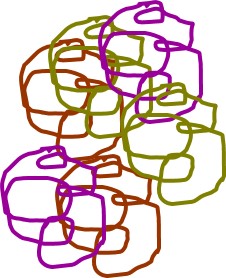
It is possible to sequence mixed DNA, but that is expensive and generally will only tell you the most abundant organisms present. Instead we will target one gene (16S rRNA gene) that is useful for taxonomic identification. This targeting will make thousands of copies of that one gene. In addition to making lots of copies of the 16S genes in our sample, we will also attach a few extra DNA bases that act like a barcode to tell us what sample that sequence came from. In my cartoon, the little blue box is the barcode for that sample.
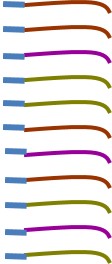
Those few bases that we attach allow us to mix together lots of samples and sequence them on one run which makes sequencing affordable.

Our samples will be sequenced using MiSeq kits provided by Illumina.
So now we have sequences. A lot of sequences. 10 million sequences. What do we do with gigabites of DNA sequences?
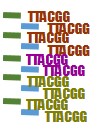
Remember those extra “barcode” bases we added when we were making copies of the 16S gene? We can use those barcode sequences to sort which sequences belong to each sample.

Once we have all the sequences sorted by which sample they belong to we have to use the information in the sequences to figure out what Bacteria they are related to. This is easier said than done because we only know about a fraction of the bacterial species that are out there. But that’s also what makes microbes so much fun to work on!
Here’s the whole workflow-IF everything works the first time for a batch of samples, this process takes about a month!