About Us
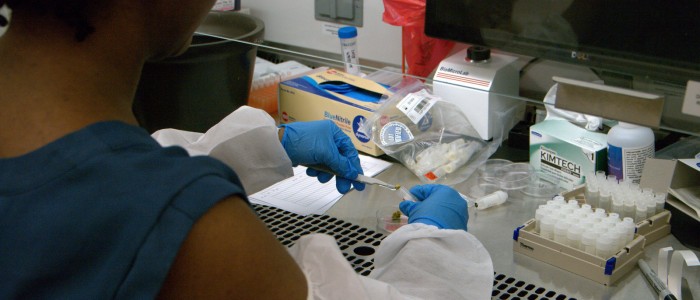
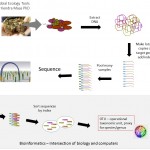
The Microbial Analysis, Resources and Services (MARS) facility supports the research community of the University of Connecticut specializing in the analysis of microbial samples and high-throughput processing of nucleic acids. Examples include the characterization of microbiomes (Bacterial V4, Bacterial EMP, Fungal ITS, or custom amplicon), sequencing of small genomes, 96-well and 384-well PCR setup or DNA quantification and other automated liquid handling applications. Services are available a la carte, ranging from fee-for-service to unassisted use of the equipment by trained and certified users.
MARS News
MARS is a certified Oxford Nanopore service lab! Contact us to discuss your long read sequencing project! Stay tuned for more information in the new year about our custom amplicon service. We are currently testing full 16S primers and will have a no-sample-minimum service next year!
We've updated out Facility Statement (see below). Please let Kendra know if you are applying for a grant that uses MARS equipment and/or services even if you don't need a letter of support.
MARS is looking for a full time AAUP technician position. Please apply here if you love bench work and want the challenge of being at the leading edge of developing methods for examining microbial communities!
We are delighted to welcome Dr. Lisa Nigro as MARS postdoctoral facility scientist. Lisa brings years of experience exploring microbial communities from a wide variety of environments.
MARS has been busy developing low cost, high throughput surveillance screens for the UConn community.
Monitoring Covid in wastewater across UConn.
COVID-19 UPDATE 2020/05/20 MARS is resuming in person lab work. We will process samples that we had received before the ramp down first. All labs wanting to resume in person lab work must apply for and receive OVPR approval. Feel free to contact us if you are planning on submitting samples while waiting for OVPR approval.
COVID-19 UPDATE 2020/05/12 MARS is working on protocols for ramping back up. See the OVPR page for guidance.
COVID-19 UPDATE 2020/03/18 Research activities are ramping down at UConn this week. Paulina and Bailey have been working extra hard this week to get as many samples processed as possible. We will have one last sequencing run Friday and will be sharing that data next week. Kendra will be available for bioinformatic and general data management assistance via video chatting during the shut down. Don't have a lab database but have been thinking about building one? Talk to Kendra, she loves databases!
2020/03/13 We continue to follow guidance from UConn. Our goal is to remain open as long as the university is allowing research. We have ~2-3 weeks worth of samples in hand that we will continue processing. If you have samples for us, there's no need to rush them over today because we won't be able to work on them for a couple of weeks. If you want to reserve your place in the queue, fill out the sample submission and we will contact you when we are ready to process your samples. We will work out sample transfer at that time. As always, reach out to us with questions . Like you, we are trying to figure out how to work or seclude as the situation develops.
MARS will be closed Dec 23-27. Happy Holidays!
Kendra is running a week long workshop on microbiome/amplicon analysis using mothur and R again, Dec 16-20. Participation limited to 15. If you have taken this workshop before and are interested in helping teach it, please let us know.
MARS welcomes our new tech Paulina!
MARS rates have changed as of the beginning of the fiscal year (July 1). See services page for details.
We have cleared out the backlog from Kendra's absence! Turn around times should be back to normal (see services page)
MARS welcomes our new tech Bailey!
Kendra is away from Mars till Feb 12 to spend time with a new baby! Response and processing times are longer than typical during this time.
Kendra is running a week long workshop on microbiome/amplicon analysis using mothur and R again, Dec 17-21. Participation limited to 15.
Kendra is offering a new short workshop. 2 day introduction to R and microbiome/ecology statistics. Aug 20-21, 2018
MARS welcomes our new tech Nicholas Hellmann to the team!
MARS has moved to the new Engineering and Sciences Building Rm 309.
Join us at CT Bioblitz 2017 in Greenwich. Kendra will have interactive data from last year's bioblitz.
Kendra is running a week long workshop on microbiome/amplicon analysis using mothur and R again this summer, Aug 21-25, 2017. Participation limited to 10.
MARS welcomes our new tech, Samantha Sanford! We wish Peter Cardoz well in his new position at Jax.
Join us for a 2 day workshop, Introduction to data visualization using R, Jan 12-13, 2017
Subscribe to our listserv for updates about MARS workshops, seminars, and instrument updates. mars-l@uconn.edu
MARS has moved to Beach Hall Rm 201. Come visit our new space!
Join us for our first week long workshop on microbiome/amplicon analysis using mothur and R, Aug 15-19, 2016. Participation limited to 10.
MARS rates have changed as of July 1
MARS is participating in the 2016 Bioblitz!
The White House issues call to action on microbiome research.
Facility Statement
The Microbial Analyses, Resources, and Services (MARS) facility within the Center for Open Research Resources and Equipment at the University of Connecticut is a specialized next generation sequencing laboratory. MARS is a service lab focused on microbial community, amplicon, and small genome sequencing as well providing the UConn research community access to our fully equipped molecular laboratory. We offer both Illumina (MiSeq) and Oxford Nanopore (Gridion) DNA/RNA sequencing platforms. Equipment available for use includes sample preparation robots: Kingfisher Apex, Eppendorf epMotion 5075 TMX and Opentron OT-2 liquid handling robots (single and 8 channel pipetting from 0.1ul 1000ul), Qiagen QiCube and QiAgility. Additional equipment: Synergy HT plate reader (fluorescence, luminescence, and absorbance), Quibit 4.0, QiAxcel nucleic acid fragment analyzer, BioRad 384 well qpcr, Covaris M220 ultra-sonicator, along with standard molecular biological equipment (thermal cyclers, centrifuges, single and multichannel micropipettors, water baths, incubators, refrigerators and freezers for sample and reagent storage). The sample processing laboratory also contains a dedicated room with a biosafety hood for handling raw samples up to BSL2 which is primarily used to prepare samples for DNA/RNA extraction. MARS offers training on all instrumentation for users interested in unsupervised use of any equipment as well as fee-for-service sample processing. Fee for service sample processing is both our custom microbiome libraries and most commercially available library kits. MARS offers bioinformatic and statistical support for microbiome analysis as workshops to train users as well as fee-for-service custom analyses.
Contact Us
MARS
Center for Open Research Resources and Equipment
Engineering and Science Building, Rm 309
181 Auditorium Rd Unit 3032
Storrs, CT 06269
Phone: 860-486-1417
E-mail: mars@uconn.edu
Dr. Kendra Maas, Facility Scientist
Follow us on Twitter @UconnMars
MARS Blog
Follow MARS on Twitter
No feed found with the ID 1. Go to the All Feeds page and select an ID from an existing feed.