If you are interested in leveraging Next Gen sequencing power but the services listed don’t exactly match what you want to do, please contact us . We’re happy to discuss custom sequencing projects including protocol development.
Turnaround time for our services is dependent on type of service and the number of samples that you submit. Our prices are based on multiplexing many samples on each run, therefore if you submit a few samples they won’t be processed till the run is filled. If you submit full runs of samples (95+ samples) for microbiome sequencing, they are usually completed in 4-8 weeks depending on our current queue. If your project is different enough from our usual process that we deem it “protocol development”, we won’t be able to estimate turn around but will have regular conversations updating you on the progress. Please contact us for current turn around time estimates.
Users that are more concerned with turnaround time than cost, please talk to us. It is often possible for your samples to be sequenced quickly on less than full runs, you would just need to pay the balance of the run cost. Our fastest turnaround is for user generated libraries and 2-step amplicon libraries, many times we return data for these types of samples in less than 2 weeks.
If you are shipping samples to us, please consult our shipping guide and/or contact us with any shipping questions.
Microbiome and Amplicon Analysis
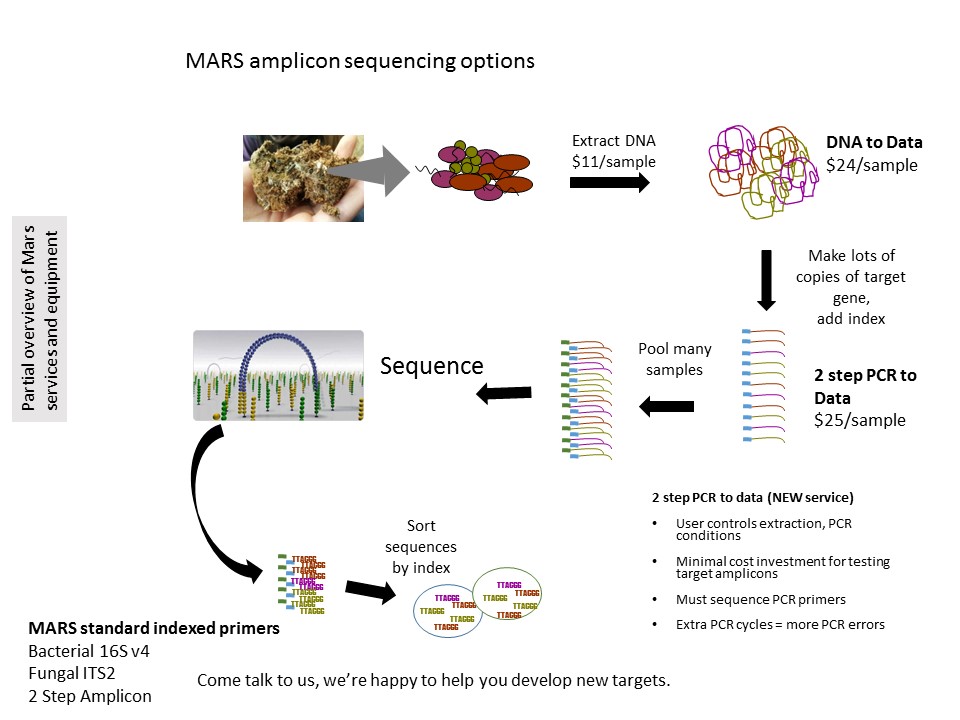
MARS offers several on-ramps for this analysis. Full service sample processing is available by combining DNA extraction with our amplicon library prep and amplicon sequencing which includes quantification, library prep, and sequencing at least 10,000 sequences per sample.
If you are interested in having us extract your samples, be aware that we use a 96 well kit optimized for soil and we often extract high biomass samples (fecal and soil are common). Especially if you are working with low biomass samples, you are going to have better luck extracting them with a method that is tuned to your sample type.
We don’t have a hard cutoff for quantity or quality of DNA, but be aware the very dilute or impure DNA may lead to difficulty amplifying and biased community recovered. If you submit DNA, we recommend at least 50ng in 20ul.
Users who either want to control all of the library prep or who are investigating multiple targets, may want to use the 2-step Amplicon to data approach where the user does the initial PCR then we attach the indexes in a second PCR. We can also sequence user generated libraries provided that they can use either MARS or Illumina standard sequencing primers (Client Amplicon library to data). If you are interested in either Client Amplicon library to data or 2-step Amplicon to data, please contact us *before* you do any library prep.
We are very mindful of limited budgets for research so aim to provide our services as cost effectively as possible. To that end, we will attempt to generate amplicon libraries 2 times varying DNA template amounts and/or additives. We typically use 10ng DNA in 15ul PCR reactions with BSA to help with inhibitors as our starting condition. If you use other conditions for PCRing your samples, please let us know! If a sample does not generate enough library after 2 tries we will contact you to determine next steps-increase PCR cycles to 35, drastically increase or decrease template amount, or sequence one of the first attempts. Our methods are summarized here.
*Notice new rates for FY22 effective for all samples processed after July 1,2021*
| Service | Internal Rate | External nonprofit Rate |
|---|---|---|
| DNA Extraction | $21/sample | $29/sample |
| Amplicon Library Prep | $11/sample | $15/sample |
| 2-step Library Prep | $7.50/sample | $11/sample |
| Client Amplicon library qc and pooling | $4/sample | $6/sample |
| Amplicon Sequencing | $16.50/sample | $21/sample |
| OTU clustering, mothur standard process | $53/batch | $74/batch |
If you would like us to cluster your data into OTUs we will use a tailored variation of this mothur processing script. The data returned will be OTU tables and alpha/beta diversity calculated for 3% OTUs for Bacteria 16S v4 or 5% for Fungal ITS2. I’ll also taxonomically ID the OTUs to genus because that’s as far as I think you can reliably identify 16S v4 data. There are data pipelines that will return species level names but I believe that says more about what sequences are in the database than what sequences are in your data. Please talk to me if you think your dataset would be appropriate for species level identification and we can decide together a course of action.
Additional custom data analysis is available including multivariate statistical analyses and publication quality figure generation, contact us for a quote, example set of analyses.
qPCR
Sequencing data does not give absolute abundance of organisms, to get absolute abundance you need to use qPCR. Usually when running qPCR you compare your sample to a known standard. MARS has standards for 16S for Bacteria and ITS2 for Fungi. If you are interested in other targets contact us and we will talk to you about designing an assay. We run samples and standards in triplicate, so can fit up to 120 samples per plate. Your cost is plate setup + Number of samples * per sample fee.
| Service | Internal Rate | External nonprofit Rate |
|---|---|---|
| qPCR plate setup | $48/plate | $65/plate |
| qPCR sample fee | $3.50/sample | $5/sample |
Genome Analysis
For BSL1 or BSL2 isolates, you can give us a cell pellet for DNA extraction before DNA to Genome processing. We also accept DNA or user generated libraries with no BSL restriction. These prices are for NexteraXT library prep which works for many but not all bacterial genomes (high GC organisms don’t work well with this kit). The data will be >300MB of 2x250bp PE reads. If you’re not sure what library prep your genome will need or if you need more or less reads contact us to discuss your project.
| Service | Internal Rate | External nonprofit Rate |
|---|---|---|
| Genome library prep Nextera XT | $55/sample | $75/sample |
| Client library to Genome | $126/sample | $170/sample |
Consultation and Education
| Service | Internal Rate | External nonprofit Rate | ||||||
|---|---|---|---|---|---|---|---|---|
| SARS-CoV2 monitoring | Internal Rate | External nonprofit Rate |
|---|---|---|
| sample fee | $36/sample | $75/sample |
Instrument Use
MARS instrumentation can be accessed either unassisted by authorize users or with the assistance of MARS personnel. Once a user has been trained on an instrument, fill out the Equipment request form to reserve time. Instrument access is billed per whole hour. Aliquots of many of the reagents and kits that MARS uses can be purchased for cost +5% aliquot fee. Contact us to find out if we have a particular reagent or kit.
| Instrument | Assisted Rate | Unassisted Rate | External Assisted Rate |
|---|---|---|---|
| MiSeq | $180/run | $150/run | $250/run |
| EpMotion 5075 | $50/hour | $20/hour | $165/hour |
| Synergy Plate Reader | $37/hour | $5/hour | $50/hour |
| Qubit | $35/hour | $5/hour | $50/hour |
| QIAxcel | Consumables and machine time for fastDNA are $7/12 sample set | $10/12 sample set | |
| QIAcube | $21/run | $55/run | |